Machine Learning Course
Trener
Krzysztof Mędrela
tel.: +48 660 873 898
email: krzysztof@medrela.com
web: www.medrela.com
Installation¶
Create universe.py File¶
Create universe.py file and put the following code there:
# %%writefile universe.py
# Ignore warning
import warnings
warnings.filterwarnings('ignore', message="numpy.dtype size changed")
warnings.filterwarnings('ignore', message="Objective did not converge")
warnings.filterwarnings('ignore', message=r"The \*bottom\* kwarg to \`barh\` is deprecated use \*y\* instead\.")
warnings.filterwarnings('ignore', message=r".*Falling back to \'gelss\' driver")
warnings.filterwarnings('ignore', message=r"Variables are collinear")
warnings.filterwarnings('ignore', message="Precision is ill-defined")
warnings.filterwarnings('ignore', message="invalid value encountered in double_scalars")
warnings.filterwarnings('ignore', message="F-score is ill-defined")
warnings.filterwarnings('ignore', message="Precision and F-score are ill-defined")
warnings.filterwarnings('ignore', message="The 'categorical_features' keyword is deprecated in version")
warnings.filterwarnings('ignore', message="posx and posy should be finite values")
# warnings.filterwarnings('ignore', category=DeprecationWarning)
# warnings.filterwarnings('ignore', category=FutureWarning)
# warnings.filterwarnings("ignore", message="numpy.core.umath_tests is an internal NumPy module")
# Builtin Libraries
from collections import defaultdict
from datetime import date
from datetime import datetime
from itertools import chain, combinations
from optparse import OptionParser
import pickle
import sys
# Third-party backports of builtin libraries
# from dataclasses import dataclass
# Third-party Libraries
# import graphviz
import matplotlib as mpl
from matplotlib import pyplot as plt
import mglearn
import numpy as np
import pandas as pd
from scipy.cluster.hierarchy import dendrogram, ward
import scipy.linalg
# Ignore some warnings from numpy
np.seterr(divide='ignore', invalid='ignore')
# Datasets Builtin Into scikit-learn
from mglearn.datasets import make_blobs as mglearn_make_blobs
from sklearn.datasets import load_boston
from sklearn.datasets import load_breast_cancer
from sklearn.datasets import load_digits
from sklearn.datasets import load_iris
from sklearn.datasets import make_blobs
from sklearn.datasets import make_moons
# Utilities from scikit-learn
from sklearn.cross_decomposition import PLSRegression
from sklearn.decomposition import PCA
from sklearn.decomposition import NMF
from sklearn.feature_selection import RFE
from sklearn.feature_selection import SelectFromModel
from sklearn.feature_selection import SelectPercentile
from sklearn.feature_selection import SelectKBest
from sklearn.manifold import TSNE
from sklearn.model_selection import cross_val_score
from sklearn.model_selection import GridSearchCV
from sklearn.model_selection import GroupKFold
from sklearn.model_selection import KFold
from sklearn.model_selection import LeaveOneOut
from sklearn.model_selection import PredefinedSplit
from sklearn.model_selection import ShuffleSplit
from sklearn.model_selection import StratifiedKFold
from sklearn.model_selection import StratifiedShuffleSplit
from sklearn.model_selection import TimeSeriesSplit
from sklearn.model_selection import train_test_split
from sklearn.pipeline import Pipeline
from sklearn.preprocessing import MinMaxScaler
from sklearn.preprocessing import OneHotEncoder
from sklearn.preprocessing import PolynomialFeatures
from sklearn.preprocessing import StandardScaler
from sklearn.preprocessing import FunctionTransformer
# Metrics
from sklearn.metrics import accuracy_score
from sklearn.metrics import average_precision_score
from sklearn.metrics import classification_report
from sklearn.metrics import cohen_kappa_score
from sklearn.metrics import confusion_matrix
from sklearn.metrics import f1_score
from sklearn.metrics import fbeta_score
from sklearn.metrics import make_scorer
from sklearn.metrics import precision_recall_curve
from sklearn.metrics import roc_auc_score
from sklearn.metrics import roc_curve
from sklearn.metrics.cluster import adjusted_rand_score
# Models from scikit-learn
from sklearn.cluster import AgglomerativeClustering
from sklearn.cluster import DBSCAN
from sklearn.cluster import KMeans
from sklearn.discriminant_analysis import LinearDiscriminantAnalysis
from sklearn.discriminant_analysis import QuadraticDiscriminantAnalysis
from sklearn.dummy import DummyClassifier
from sklearn.ensemble import BaggingRegressor
from sklearn.ensemble import GradientBoostingClassifier
from sklearn.ensemble import GradientBoostingRegressor
from sklearn.ensemble import RandomForestClassifier
from sklearn.ensemble import RandomForestRegressor
from sklearn.linear_model import ElasticNet
from sklearn.linear_model import Lasso
from sklearn.linear_model import LinearRegression
from sklearn.linear_model import LogisticRegression
from sklearn.linear_model import Ridge
from sklearn.naive_bayes import GaussianNB
from sklearn.naive_bayes import MultinomialNB
from sklearn.neighbors import KNeighborsClassifier
from sklearn.neighbors import KNeighborsRegressor
from sklearn.neural_network import MLPClassifier
from sklearn.svm import SVC
from sklearn.tree import DecisionTreeClassifier
from sklearn.tree import DecisionTreeRegressor
from sklearn.tree import export_graphviz
# imbalanced-learn
#from imblearn.under_sampling import RandomUnderSampler
#from imblearn.over_sampling import RandomOverSampler
# Deep Learning
# import keras
# from keras.datasets import boston_housing
# from keras.datasets import cifar10
# from keras.datasets import mnist
# from keras.layers import Conv2D
# from keras.layers import Dense
# from keras.layers import Dropout
# from keras.layers import Flatten
# from keras.layers import LSTM
# from keras.layers import MaxPooling2D
# from keras.models import load_model
# from keras.models import Sequential
# from keras.preprocessing.image import ImageDataGenerator
# from keras.utils import to_categorical
# import tensorflow
# Finish
python_version = sys.version.split(" ")[0]
print(f"Python version: {python_version}")
print("Universe has been successfully imported.")
from universe import *
Data Visualization with matplotlib¶
%matplotlib inline
from matplotlib import pyplot as plt
Basic Line Plots¶
Y¶
plt.figure(figsize=(10, 5))
plt.plot([1, 3, 2])
plt.show()
X and Y¶
plt.figure(figsize=(10, 5))
x = [5.0, 12.5, 8.0]
y = [1, 3, 2]
plt.plot(x, y)
plt.show()
Custom Area¶
plt.figure(figsize=(10, 5))
plt.plot([1, 3, 2])
plt.xlim(-1, 3) # Setter
plt.ylim(-5, 5)
print(plt.xlim()) # Getter
plt.show()
Grid¶
plt.figure(figsize=(10, 3))
plt.plot([1, 3, 2])
plt.grid()
plt.show()
Multiple Series¶
plt.figure(figsize=(10, 5))
plt.plot([1, 3, 2]) # First Series
plt.plot([5, 4, 1.5]) # Second Series
plt.show()
Legend¶
plt.figure(figsize=(10, 5))
plt.plot([1, 3, 2], label='A')
plt.plot([5, 4, 1.5])
plt.legend() # loc='lower left'
plt.show()
Texts¶
plt.figure(figsize=(10, 5))
plt.plot([5.0, 5.5, 7.0], [1, 3, 2])
plt.title('Title')
plt.xlabel('xlabel')
plt.ylabel('ylabel')
plt.show()
Custom Line Style¶
plt.figure(figsize=(10, 5))
# Hit shift+Tab for more options
plt.plot([1, 3, 2], 'r--') # r=red
plt.plot([5, 4, 1.5], 'gx') # g=green
plt.show()
# Hit Shift+Tab for more customization options.
Exercise: Basic Line Plots¶
### Data
import numpy as np
from matplotlib import pyplot as plt
# Generating Data
x = np.arange(0.0, 10.0, 0.1)
y1 = np.exp(-x)
y2 = np.exp(-x) * np.cos(2*np.pi*x)
# Printing Data
print(f"x[:7] = {x[:7]}")
print(f"y1[:4] = {y1[:4]}")
print(f"y2[:4] = {y2[:4]}")
### Your Code Here
# y - yellow
# b - blue
# o - circle
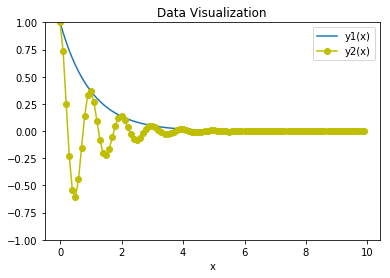
### Solution
plt.figure()
plt.plot(x, y1, label="y1(x)")
plt.plot(x, y2, 'yo-', label="y2(x)")
plt.xlabel("x")
plt.ylim(-1, 1)
plt.legend()
plt.title("Data Visualization")
plt.show()
Scales¶
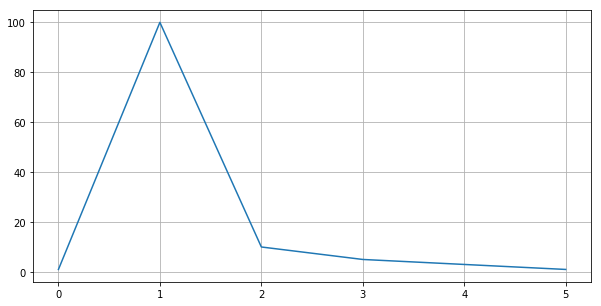
Linear Scale¶
plt.figure(figsize=(10, 5))
plt.plot([1, 100, 10, 5, 3, 1])
# plt.yscale('log') # 'linear', log', 'symlog', 'logit'
plt.grid()
plt.show()
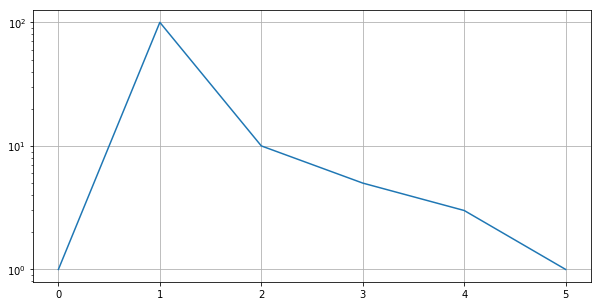
Log Scale¶
plt.figure(figsize=(10, 5))
plt.plot([1, 100, 10, 5, 3, 1])
plt.yscale('log') # 'linear', log', 'symlog', 'logit'
plt.grid()
plt.show()
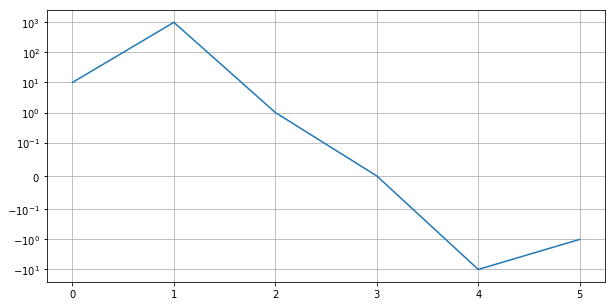
Symlog Scale¶
plt.figure(figsize=(10, 5))
plt.plot([10, 1000, 1, 0, -10, -1])
plt.yscale('symlog', linthreshy=0.1)
plt.grid()
plt.show()
Logit Scale¶
plt.figure(figsize=(8, 4))
plt.plot([0.1, 0.9, 0.99, 0.999])
plt.yscale('logit')
plt.show()
Exercise: Scales¶
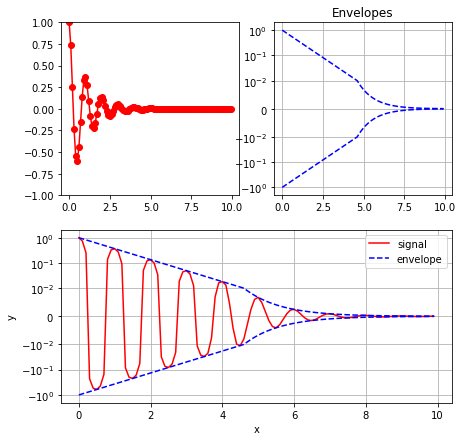
### Solution
plt.figure(figsize=(6,6))
plt.yscale('symlog', linthreshy=0.01)
plt.plot(x, y1, 'b--')
plt.plot(x, -y1, 'b--')
plt.title("Envelopes")
plt.grid()
Multiple Plots¶
Subplot¶
plt.figure(figsize=(10, 5))
plt.subplot(1, 2, 1)
plt.plot([1, 3, 2])
plt.subplot(1, 2, 2)
plt.plot([5, 4, 1.5])
plt.show()
More Advanced Subplot¶
plt.figure(figsize=(10, 5))
plt.subplot(1, 2, 1) # left half
plt.plot([1, 3, 2])
plt.subplot(2, 2, 2) # right top
plt.plot([5, 4, 1.5])
plt.subplot(2, 2, 4) # right bottom
plt.plot([5, 4, 5.5])
plt.show()
Exercise: Multiple Plots¶
### Data
import numpy as np
from matplotlib import pyplot as plt
# Generating Data
x = np.arange(0.0, 10.0, 0.1)
y1 = np.exp(-x)
y2 = np.exp(-x) * np.cos(2*np.pi*x)
# Printing Data
print(f"x[:7] = {x[:7]}")
print(f"y1[:4] = {y1[:4]}")
print(f"y2[:4] = {y2[:4]}")
# Your Code Here
### Solution
plt.figure(figsize=(7, 7))
plt.subplot(2, 2, 1)
plt.plot(x, y2, 'ro-')
plt.ylim(-1, 1)
plt.subplot(2, 2, 2)
plt.yscale('symlog', linthreshy=0.01)
plt.plot(x, y1, 'b--')
plt.plot(x, -y1, 'b--')
plt.title("Envelopes")
plt.grid()
plt.subplot(2, 1, 2)
plt.yscale('symlog', linthreshy=0.01)
plt.plot(x, y2, 'r', label="signal")
plt.plot(x, y1, 'b--', label="envelope")
plt.plot(x, -y1, 'b--')
plt.xlabel("x")
plt.ylabel("y")
plt.legend()
plt.grid()
plt.show()
Introduction to Machine Learning¶
from universe import *
What is Machine Learning?¶

Artificial Intelligence -- intelligence demonstrated by machines, in contrast to the natural intelligence displayed by humans and other animals.
Machine Learning -- part of AI, a field of computer science that uses statistical techniques to give computer systems the ability to "learn" (e.g., progressively improve performance on a specific task) with data, without being explicitly programmed.
Deep Learning -- part of Machine Learning, part of a broader family of machine learning methods based on learning data representations, as opposed to task-specific algorithms. To cut the long story short, it's all about neural networks.
Basic Concepts¶
Petal vs sepal¶

Input: petal and sepal lengths and widths.
Multiclass output: setosa, versicolor, or virginica.
Data¶

Basic Concepts ¶
- Dataset = a table of data
- Feature = one input column
- Featureset = all input columns
- Sample, data point, observation = one row
- Model or algorithm = a black box
- Input data, input, featureset, X = all input columns
- Output data, output, Y, target = the column that we want to predict
- Feature engineering and selection = choosing the best data representation
- Training and test data/set
- Supervised Learning = learning with the ground truth
- Unsupervised Learning = learning without ground truth
- Data type = either continuous (float or integer) or a category (binary or integer)
Case Study: Iris Classification¶
Step 1: Gather Data¶
Nothing to do. :-)
Step 2: Load Data¶
from universe import *
iris_dataset = load_iris()
X = iris_dataset['data']
y = iris_dataset['target']
np.random.seed(0)
y[::3] = np.random.randint(0, 3, size=(y.size//3)) # add some noise
print(X.shape)
print(iris_dataset['feature_names'])
print(iris_dataset['target_names'])
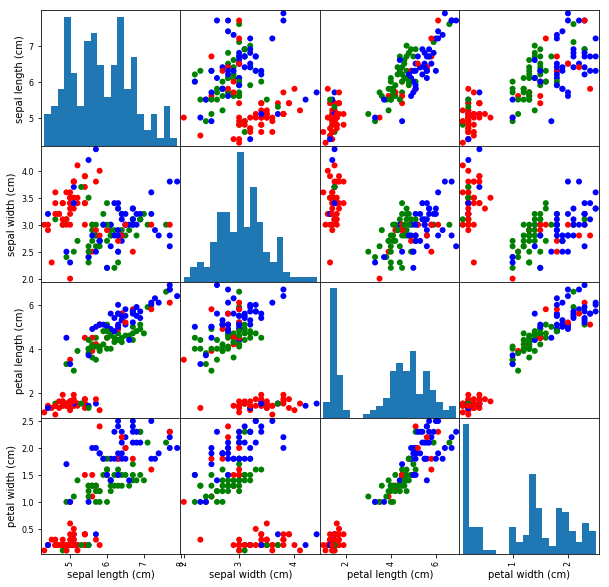
Step 3: Explore & Visualise Data¶
print(X[:5])
print(y)
X_df = pd.DataFrame(X, columns=iris_dataset['feature_names'])
X_df['category'] = y
display(X_df[:5])
X_df = pd.DataFrame(X, columns=iris_dataset['feature_names'])
pd.plotting.scatter_matrix(
X_df,
c=y,
figsize=(10, 10),
marker='o',
hist_kwds={'bins': 20},
alpha=1.0,
cmap=mpl.colors.ListedColormap(['r', 'g', 'b']),
);
Step 4: Clean Data¶
Nothing to do. :-)
Step 5: Choose Data Representation¶
Let's stick to the original featureset.
Step 6: Build and Train Model¶
Step 6a: Train and Test Datasets¶
# Split dataset into train and test sets
X_train, X_test, y_train, y_test = train_test_split(
X, y, random_state=0)
print(f"X_train.shape = {X_train.shape}")
print(f"X_test.shape = {X_test.shape}")
print(f"y_train.shape = {y_train.shape}")
print(f"y_test.shape = {y_test.shape}")
print(y_test)
Step 6b: Build and Train Model¶
knn = KNeighborsClassifier(n_neighbors=1) # Build Model
knn.fit(X_train, y_train); # Train Model
Step 7: Evaluate Model¶
y_pred = knn.predict(X_test)
print(y_pred)
print(np.mean(y_pred == y_test))
print(knn.score(X_test, y_test)) # Exactly the same as above
print(knn.score(X_train, y_train))
Complete Script¶
from universe import *
# Step 1: Gather Data
# nothing to do
# Step 2: Load Data
iris_dataset = load_iris()
X = iris_dataset['data']
y = iris_dataset['target']
np.random.seed(0)
y[::3] = np.random.randint(0, 3, size=(y.size//3)) # add some noise
# Step 3: Visualize and Explore Data
X_df = pd.DataFrame(X, columns=iris_dataset['feature_names'])
pd.plotting.scatter_matrix(
X_df,
c=y,
figsize=(10, 10),
marker='o',
hist_kwds={'bins': 20},
alpha=1.0,
cmap=mpl.colors.ListedColormap(['r', 'g', 'b']),
)
# Step 4: Clean Data
# nothing to do
# Step 5: Choose Data Representation
# Step 5a: Data Preprocessing (i.e. Normalization)
# Step 5b: Feature Engineering
# Step 5c: Feature Selection
# nothing to do
# Step 6: Build and Train Model
# Split dataset into train and test sets
X_train, X_test, y_train, y_test = train_test_split(
X, y, random_state=0)
knn = KNeighborsClassifier(n_neighbors=1) # Build Model
knn.fit(X_train, y_train) # Train Model
# Step 7: Evaluate Model
prediction = knn.predict(np.array([[5, 2.9, 1 ,0.2]]))
train_score = knn.score(X_train, y_train)
test_score = knn.score(X_test, y_test)
print(f"TRAINING SCORE: {train_score:.3f}")
print(f"TEST SCORE: {test_score:.3f}")
Common Workflow¶

- Gathering Data
- Loading Data
- Data Exploration and Visualization
- Cleaning
- Choosing Data Representation
- Data Preprocessing (i.e. Normalization)
- Feature Engineering
- Feature Selection
- Building and Training a Model
- Evaluating the Model
Problem Types¶

Supervised Learning¶

Bias-Variance Trade Off¶
Underfitting and Overfitting¶
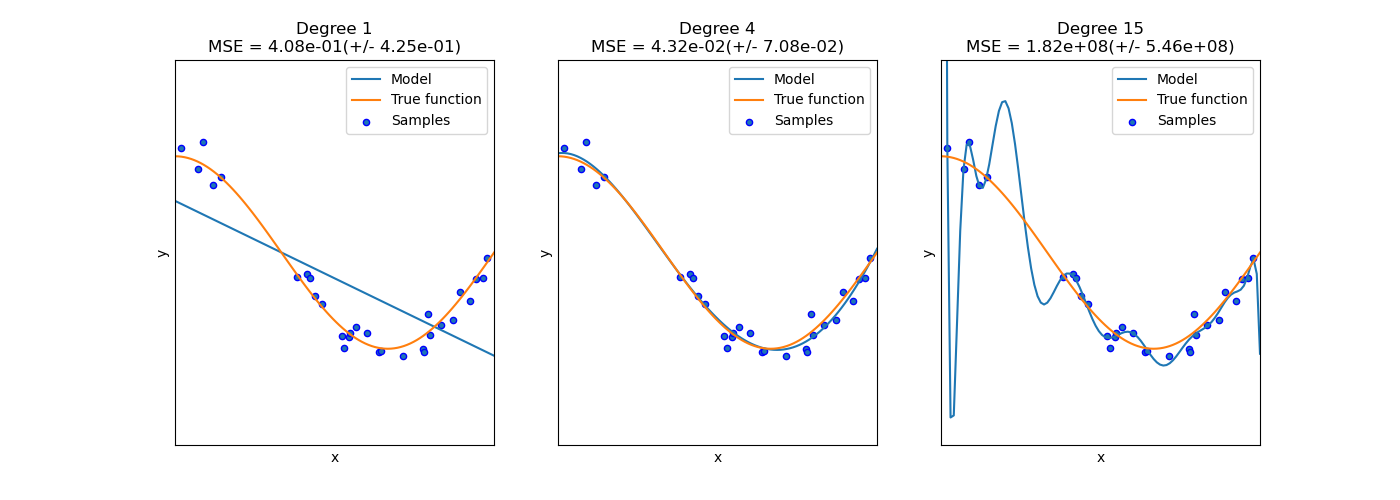
Bias and Variance¶
- The bias is an error from erroneous assumptions in the learning algorithm. High bias can cause an algorithm to miss the relevant relations between features and target outputs (underfitting).
- The variance is an error from sensitivity to small fluctuations in the training set. High variance can cause an algorithm to model the random noise in the training data, rather than the intended outputs (overfitting).
Sweet Spot¶

KNN Complexity¶
Iris Dataset¶
Resources¶
- Book: Introduction to Machine Learning with Python by Andreas C. Muller along with code snippets
- Scikit-learn Documentation (User Guide)
- Kaggle.com - data science projects
Regression Linear Models¶
from universe import *
%matplotlib inline
Simple Linear Regression¶
Model & Cost Function¶
Model:
$$ y_\textit{prediction} = w \cdot x + b$$
Cost function (Ordinary Least Squares):
$$ \underset{w, b}{min\,} \frac{1}{2n_{samples}} \sum\limits_{i}^{\text{samples}} (y_\textit{prediction} - y_\textit{ground truth})^2 $$
Example Model¶
from universe import *
# Load and Split Data
X, y = mglearn.datasets.make_wave(n_samples=60)
y += 1.5
X_train, X_test, y_train, y_test = train_test_split(X, y, random_state=42)
# Build and Train Model
model = LinearRegression()
model.fit(X_train, y_train);
The Dataset¶
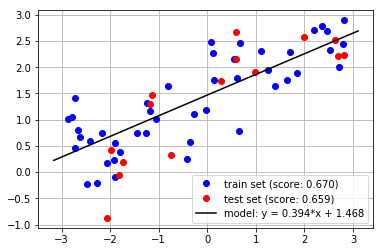
### Solution
# Evaluate Model
train_score = model.score(X_train, y_train)
test_score = model.score(X_test, y_test)
# Plot Figure
plt.figure()
plt.plot(X_train, y_train, 'bo',
label=f"train set (score: {train_score:.3f})")
plt.plot(X_test, y_test, 'ro',
label=f"test set (score: {test_score:.3f})")
# xx = np.linspace(*plt.xlim()).reshape(-1, 1)
xx = np.array(plt.xlim()).reshape(-1, 1)
yy = model.predict(xx)
plt.plot(xx, yy, 'k',
label=f"model: y = {model.coef_[0]:.3f}*x "
f"+ {model.intercept_:.3f}")
plt.legend()
plt.grid()
plt.show()
Correlation¶
correlation = np.corrcoef(X_train.reshape(-1), y_train)[0, 1]
print(f"CORRELATION: {correlation:.3f}")
print(f"COEFFICIENTS: {model.coef_}")
print(f"INTERCEPT: {model.intercept_}")

Multiple Linear Regression¶
Model & Cost Function¶
Model ($k$ features):
$$ y_\textit{prediction} = w_1 \cdot x_1 + \dots + w_k \cdot x_k + b$$
Cost function (Ordinary Least Squares):
$$ \underset{w_1,\dots,w_k,b}{min\,} \frac{1}{2n_{samples}} \sum\limits_{i}^{\text{samples}} (y_\textit{prediction} - y_\textit{ground truth})^2 $$
Boston Dataset¶
Boston dataset has 13 features.
The output is median value of owner-occupied homes in $1000's.
How do you explore dataset with so many features?
Features:
- CRIM - per capita crime rate by town
- ZN - proportion of residential land zoned for lots over 25,000 sq.ft.
- INDUS - proportion of non-retail business acres per town.
- CHAS - Charles River dummy variable (1 if tract bounds river; 0 otherwise)
- NOX - nitric oxides concentration (parts per 10 million)
- RM - average number of rooms per dwelling
- AGE - proportion of owner-occupied units built prior to 1940
- DIS - weighted distances to five Boston employment centres
- RAD - index of accessibility to radial highways
- TAX - full-value property-tax rate per \$10,000
- PTRATIO - pupil-teacher ratio by town
- B - 1000(Bk - 0.63)^2 where Bk is the proportion of blacks by town
- LSTAT - % lower status of the population
from universe import *
dataset = load_boston()
X, y = dataset['data'], dataset['target']
feature_names = dataset['feature_names']
Explore Dataset¶
Xdf = pd.DataFrame(X, columns=feature_names)
display(Xdf.describe().T)
Results¶
from universe import *
# Load and Split Data
dataset = load_boston()
X, y = dataset['data'], dataset['target']
feature_names = dataset['feature_names']
X_train, X_test, y_train, y_test = train_test_split(X, y, random_state=42)
# Build and Train Model
model = LinearRegression()
model.fit(X_train, y_train)
# Print Results
print(f"INTERCEPT: {model.intercept_}")
print(f"COEFFS: {model.coef_}")
train_score = model.score(X_train, y_train)
test_score = model.score(X_test, y_test)
print(f"Train score: {train_score:.3f}")
print(f"Test score: {test_score:.3f}")
Exercise: Regression Lab¶
from universe import *
%matplotlib inline
Generate Dataset¶
from universe import *
np.random.seed(0)
# Function to Model
def f(x, noise=0.0):
y = x[0] * (1 - 4*x[1]) # xor # the same as x[0] - 4*x[0]*x[1]
if x[2] == 0:
y += 1
elif x[2] == 1:
y += 5
elif x[2] == 2:
y += 3
y += np.random.normal(scale=noise) # noise
return y
# Featureset
X = np.array([
[x0+np.random.normal(scale=0.2), x1+np.random.normal(scale=0.2), x2]
for x0 in [0, 0.33, 0.66, 1]
for x1 in [0, 0.5, 1]
for x2 in [0, 1, 2]
]) # List Comprehension
# Noisy output
y_noisy = np.apply_along_axis(f, 1, X, noise=0.3).reshape(-1, 1)
data_noisy = np.concatenate([X, y_noisy], axis=1)
df_noisy = pd.DataFrame(data_noisy, columns=['x0', 'x1', 'x2', 'y'])
df_noisy.to_csv('regression-lab-data-noisy.csv') #, index=False)
# Print Dataset
display(df_noisy[:5])
! dir
! head -3 regression-lab-data-noisy.csv
LinearRegression¶
Use LinearRegression on the dataset. Evaluate the model and print its parameters.
# Hints:
# W nowym notebooku!
from universe import *
data = pd.read_csv('regression-lab-data-noisy.csv')
cols = ['x0', 'x1', 'x2']
# X = data[cols] # selecting multiple columns
# y = data[...] # selecting one column
# ... = train_test_split(..., random_state=42)
### Solution
from universe import *
# Load Data
data = pd.read_csv('regression-lab-data-noisy.csv')
original_featureset = ['x0', 'x1', 'x2']
X = data[original_featureset]
y = data['y']
X_train, X_test, y_train, y_test = train_test_split(
X, y, random_state=42)
# Build and Fit Model
model = LinearRegression()
model.fit(X_train, y_train)
# Evaluate Model
train_score = model.score(X_train, y_train)
test_score = model.score(X_test, y_test)
# Print Results
print(f"INTERCEPT: {model.intercept_}")
print(f"COEFFICIENTS: {model.coef_}")
print(f"Train score: {train_score:.3f}")
print(f"Test score: {test_score:.3f}")
Pipelines¶
Refactor your code: put the model in a pipeline and evaluate the entire pipeline.
One-Hot Encoding¶
Use one-hot encoding for the categorical feature x2.
# Hint:
import warnings
warnings.filterwarnings('ignore') # To silence warnings
### Solution
from universe import *
import warnings
warnings.filterwarnings('ignore') # To silence warnings
# Load Data
data = pd.read_csv('regression-lab-data-noisy.csv')
original_featureset = ['x0', 'x1', 'x2']
X = data[original_featureset]
y = data['y']
X_train, X_test, y_train, y_test = train_test_split(
X, y, random_state=42)
# Fit Model
pipeline = Pipeline(steps=[
('onehot', OneHotEncoder(categorical_features=[2], sparse=False)),
('model', LinearRegression()),
])
pipeline.fit(X_train, y_train)
# Evaluate Model
train_score = pipeline.score(X_train, y_train)
test_score = pipeline.score(X_test, y_test)
# Print Results
print(f"INTERCEPT: {pipeline.named_steps['model'].intercept_}")
print(f"COEFFICIENTS: {pipeline.named_steps['model'].coef_}")
print(f"Train score: {train_score:.3f}")
print(f"Test score: {test_score:.3f}")
Polynominal Features¶
Add Feature Engineering with Polynominal Features. Guess the right value for degree parameter.
### Solution
from universe import *
import warnings
warnings.filterwarnings('ignore') # To silence warnings
# Load Data
data = pd.read_csv('regression-lab-data-noisy.csv')
original_featureset = ['x0', 'x1', 'x2']
X = data[original_featureset]
y = data['y']
X_train, X_test, y_train, y_test = train_test_split(X, y, random_state=42)
# Fit Model
pipeline = Pipeline(steps=[
('onehot', OneHotEncoder(categorical_features=[2], sparse=False)),
('poly', PolynomialFeatures(degree=3, include_bias=False)),
('model', LinearRegression()),
])
pipeline.fit(X_train, y_train)
# Evaluate Model
train_score = pipeline.score(X_train, y_train)
test_score = pipeline.score(X_test, y_test)
# Print Results
print(f"INTERCEPT: {pipeline.named_steps['model'].intercept_}")
print(f"COEFFICIENTS: {pipeline.named_steps['model'].coef_}")
print(f"Train score: {train_score:.3f}")
print(f"Test score: {test_score:.3f}")
Grid Search¶
Put the pipeline in a GridSearchCV and find optimal value for degree parameter in an automated way.
### Solution
from universe import *
import warnings
warnings.filterwarnings('ignore') # To silence warnings
# Load Data
data = pd.read_csv('regression-lab-data-noisy.csv')
original_featureset = ['x0', 'x1', 'x2']
X = data[original_featureset]
y = data['y']
X_train, X_test, y_train, y_test = train_test_split(X, y, random_state=42)
# Fit Model
pipeline = Pipeline(steps=[
('onehot', OneHotEncoder(categorical_features=[2], sparse=False)),
('poly', PolynomialFeatures(include_bias=False)),
('model', LinearRegression()),
])
param_grid = {
'poly__degree': [1, 2, 3],
}
gs = GridSearchCV(
pipeline,
param_grid,
cv=KFold(n_splits=5, random_state=42),
iid=False,
return_train_score=True,
)
gs.fit(X_train, y_train)
# Evaluate Model
test_score = gs.best_score_
final_score = gs.score(X_test, y_test)
# Print Results
print(f"INTERCEPT: {gs.best_estimator_.named_steps['model'].intercept_}")
print(f"COEFFICIENTS: {gs.best_estimator_.named_steps['model'].coef_}")
print(f"BEST PARAMS: {gs.best_params_}")
print(f"Test score: {test_score:.3f}")
print(f"Final evaluation score: {final_score:.3f}")
Model Persistence¶
Saving¶
import pickle
stream = open('model.model', 'wb')
pickle.dump(gs.best_estimator_, stream)
Loading¶
import pickle
stream = open('model.model', 'rb')
pipeline = pickle.load(stream)
pipeline.predict(X_test)
Feature Engineering & Selection¶
from universe import *
%matplotlib inline
Pipelines¶
Idea of Pipelines¶

PipelineFlow¶

Usage¶
### Hidden Cell -- just to make the usage working
X_train, y_train = np.array([[1]]), np.array([1])
pipeline = Pipeline(steps=[
# Any number of transformers
# ('poly', PolynomialFeatures()), # example transformer
# and then a model at the end of the pipeline
('model', LinearRegression()),
])
pipeline.fit(X_train, y_train)
train_score = pipeline.score(X_train, y_train)
test_score = pipeline.score(X_test, y_test)
prediction = pipeline.predict(X_train)
coefs = pipeline.named_steps['model'].coef_
intercept = pipeline.named_steps['model'].intercept_
Exercise¶
from universe import *
# Dataset
dataset = pd.DataFrame({
'group': [0, 0, 0, 1, 1, 1, 2, 2, 2],
'x': [10, 20, 30, 10, 20, 30, 10, 20, 30],
'y': [420, 440, 460, 120, 140, 160, 1020, 1040, 1060]
})
# y = 2*x + (300, 100 or 1000 depending on the group)
# Split dataset
X = dataset[['group', 'x']]
y = dataset['y']
X_train, y_train = X, y # Use all data for training => no test set
### Exercise:
# 1) Build and Train Pipeline (use LinearRegression)
# 2) Make Prediction
# 3) Evaluate Model
# 4) Print Results
# Hints:
# dataset['new_column'] = ... # Creates a new column
# ous model is predict = coef[0] * group + coef[1] * x + intercept
One Hot Encoding¶
Usage¶
features = [0] # indexes of categorical features
features = [True, False] # or a mask
pipeline = Pipeline(steps=[
('onehot', OneHotEncoder(categorical_features=features, sparse=False)),
('model', LinearRegression()),
])
Exercise¶
Add one hot encoding to the previous exercise.
# Hint
import warnings
warnings.filterwarnings('ignore') # To silence warnings
# predict_onehot = coef[group] + coef[3] * x + intercept
### Solution
pipeline = Pipeline(steps=[
('onehot', OneHotEncoder(
categorical_features=[True, False], sparse=False)),
('model', LinearRegression()),
])
pipeline.fit(X_train, y_train)
# Predict
dataset['predict_onehot'] = pipeline.predict(X_train)
# Evaluate Model
train_score = pipeline.score(X_train, y_train)
# Print Results
coefs = pipeline.named_steps['model'].coef_
intercept = pipeline.named_steps['model'].intercept_
print(f"COEFFICIENTS: {pipeline.named_steps['model'].coef_}")
print(f"INTERCEPT: {pipeline.named_steps['model'].intercept_:.0f}")
print(f"Train score: {train_score:.3f}")
display(dataset)
Polynominal & Interaction Terms¶
Idea¶
X = np.array([[1, 20], [2, 30]]) # two features
poly = PolynomialFeatures(degree=3, include_bias=False)
poly.fit(X)
feature_names = poly.get_feature_names()
print("FEATURE NAMES:")
for feature in poly.get_feature_names():
print("-", feature)
Example¶
from universe import *
# Load and Split Data
X, y = mglearn.datasets.make_wave(n_samples=60)
y += 1.5
X_train, X_test, y_train, y_test = train_test_split(X, y, random_state=0)
# Define Model and Param Space
pipeline = Pipeline(steps=[
('poly', PolynomialFeatures(degree=3, include_bias=False)),
('model', LinearRegression()),
])
# Train Model with Grid Search and Cross Validation
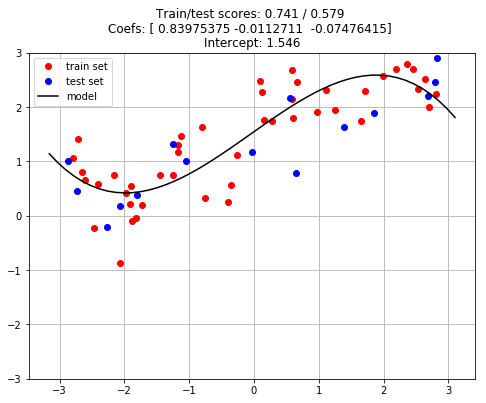
pipeline.fit(X_train, y_train);
# Evaluate
train_score = pipeline.score(X_train, y_train)
test_score = pipeline.score(X_test, y_test)
# Plot Best Estimator
plt.figure(figsize=(8, 6))
plt.plot(X_train, y_train, 'o', color='r', label='train set')
plt.plot(X_test, y_test, 'o', color='b', label='test set')
xmin, xmax = plt.xlim()
xx = np.linspace(xmin, xmax).reshape(-1, 1)
yy = pipeline.predict(xx)
plt.plot(xx, yy, 'k', label='model')
plt.legend()
plt.title(f"Train/test scores: {train_score:.3f} / {test_score:.3f} \n"
f"Coefs: {pipeline.named_steps['model'].coef_} \n"
f"Intercept: {pipeline.named_steps['model'].intercept_:.3f}")
plt.ylim([-3, 3])
plt.grid()
plt.show()
Feature Selection¶
Strategies¶
- Univariate Statistics:
SelectKBest
Toy Dataset (Noised Breast Cancer)¶
from universe import *
# Load Dataset
cancer = load_breast_cancer()
X, y = cancer['data'], cancer['target']
# Add Noise Features to the Dataset
# get deterministic random numbers
rng = np.random.RandomState(42)
noise = rng.normal(size=(len(cancer.data), 50))
# add noise features to the data
# the first 30 features are from the dataset, the next 50 are noise
X_w_noise = np.hstack([X, noise])
# Split Dataset
X_train, X_test, y_train, y_test = train_test_split(
X_w_noise, y, random_state=0, test_size=.5)
Compare Selectors¶
# Train and Evaluate Model without Feature Selection
model = LinearRegression()
model.fit(X_train, y_train)
train_score = model.score(X_train, y_train)
test_score = model.score(X_test, y_test)
print(" train / test SELECTED INFORMATIVE FEATURES")
print(f"No selection : {train_score:.3f} / {test_score:.3f} " + "X"*30 + " (30 informative + 50 noisy)")
# Define Selectors
n_features = 20
selectors = [
SelectKBest(k=n_features),
]
# Test Each Selector
for select in selectors:
# Perform Feature Selection
select.fit(X_train, y_train)
# Transform Training set
X_train_selected = select.transform(X_train)
X_test_selected = select.transform(X_test)
# Find Selected Features
support = select.get_support()[:30].astype('int')
chars = np.array([".", "X"])
selected_features = "".join(chars[support])
count = support.sum()
# Train and Evaluate Model with Feature Selection
model = LinearRegression()
model.fit(X_train_selected, y_train)
train_score = model.score(X_train_selected, y_train)
test_score = model.score(X_test_selected, y_test)
# Print Result
print(f"{select.__class__.__name__:16}: {train_score:.3f} / {test_score:.3f} {selected_features} ({count} informative + {n_features-count} noisy)")
print("")
print(f"X_train.shape: {X_train.shape}")
print(f"X_train_selected.shape: {X_train_selected.shape}")
Usage¶
pipeline = Pipeline(steps=[
# Feature Engineering
('onehot', OneHotEncoder(categorical_features=[2], sparse=False)),
('poly', PolynomialFeatures(degree=2)),
# Feature Selection
('select', SelectKBest(k=20)),
# Model
('model', LinearRegression()),
])
Inception¶
from universe import *
%matplotlib inline
Cross Validation¶
Why 80% / 20% for train/test?¶
- 99% / 1% => high variance
- 1% / 99% => high bias
- Cross Validation let us ensure that our evaluation has both low bias AND low variance.
Idea of Cross Validation¶

Cross Validation Strategies¶
k-Fold Cross Validation¶
from universe import *
X = np.array(["a", "b", "c", "d", "e", "f", "g", "h", "i"])
kf = KFold(n_splits=3)
print("Train set test set")
for train_set, test_set in kf.split(X):
print(f"{train_set} {test_set}")
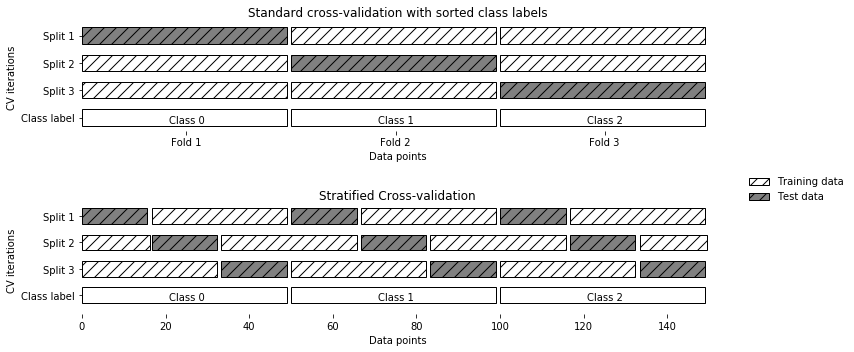
Stratified k-Fold Cross Validation¶
from universe import *
X = np.ones(10)
y = [0, 0, 0, 0, 1, 1, 1, 1, 1, 1]
skf = StratifiedKFold(n_splits=3)
print("Train set test set")
for train_set, test_set in skf.split(X, y):
print(f"{train_set} {test_set}")
Group K-Fold¶
from universe import *
X = range(10)
y = range(10)
# index: 0 1 2 3 4 5 6 7 8 9
groups = [1, 1, 1, 2, 2, 2, 3, 3, 2, 4]
print("K-FOLD")
kf = KFold(n_splits=3)
for train_set, test_set in kf.split(X, y):
print(f"{train_set} {test_set}")
print("\nGROUP K-FOLD")
gkf = GroupKFold(n_splits=3)
for train_set, test_set in gkf.split(X, y, groups=groups):
print(f"{train_set} {test_set}")
Time Series Split¶
from universe import *
X = y = range(12)
cv = TimeSeriesSplit(n_splits=3)
for train_set, test_set in cv.split(X):
print(f"{train_set} {test_set}")
Grid Search¶
Idea of Grid Search¶

Usage¶
param_grid = {
'model__n_neighbors': [1, 2, 3, 4],
'poly__degree': [1, 2, 3],
}
pipeline = Pipeline(steps=[
('poly', PolynomialFeatures(include_bias=False)),
('model', KNeighborsClassifier()),
])
gs = GridSearchCV(
pipeline,
param_grid,
cv=StratifiedKFold(n_splits=5, random_state=42),
return_train_score=True,
iid=False,
)
gs.fit(X_train, y_train);
Grid Search Outputs¶
print(f"BEST PARAMS: {gs.best_params_}")
print(f"VALIDATION SCORE: {gs.best_score_}")
n_neighbors = gs.best_params_['model__n_neighbors']
print(f"n_neighbors = {n_neighbors}")
n = gs.best_estimator_.named_steps['poly'].n_output_features_
print(f"OUTPUT FEATURES: {n}")
print(gs.best_estimator_.score(X_train, y_train) == gs.score(X_train, y_train))
GridSearch.cvresults¶
display(pd.DataFrame(gs.cv_results_)[:3].T)
Grid Search Example¶
from universe import *
# Load and Split Data
iris = load_iris()
X, y = iris['data'], iris['target']
np.random.seed(0)
y[::2] = np.random.randint(0, 3, size=(y.size//2)) # add some noise
# train set is split into train and holdout sets during cross validation
X_train, X_test, y_train, y_test = train_test_split(
X, y, random_state=0, stratify=y)
# Define Model and Param Space
param_grid = {
'model__n_neighbors': np.arange(1, 70),
}
pipeline = Pipeline(steps=[
('model', KNeighborsClassifier())
])
gs = GridSearchCV(
pipeline,
param_grid,
cv=StratifiedKFold(n_splits=5, random_state=42),
return_train_score=True,
iid=False,
)
gs.fit(X_train, y_train)
# Evaluate Model
final_score = gs.score(X_test, y_test)
# Summarize
print(f"Best params: {gs.best_params_}")
print(f"Test score: {gs.best_score_:.3f}")
print(f"Final score: {final_score:.8f}")
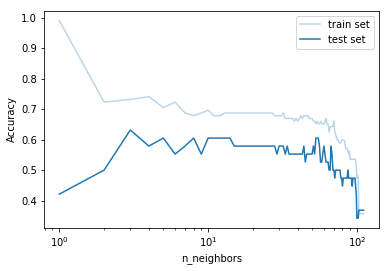
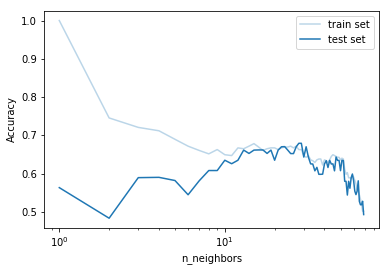
# Prepare data for the plot
k = gs.cv_results_['param_model__n_neighbors']
train_scores = gs.cv_results_['mean_train_score']
test_scores = gs.cv_results_['mean_test_score']
# Plot
plt.plot(k, train_scores, 'C0', alpha=0.3, label="train set")
plt.plot(k, test_scores, 'C0', label="test set")
plt.ylabel("Accuracy")
plt.xlabel("n_neighbors")
plt.xscale('log')
plt.legend()
plt.show()
Models¶
K-Nearest Neighbors¶
k Parameter¶
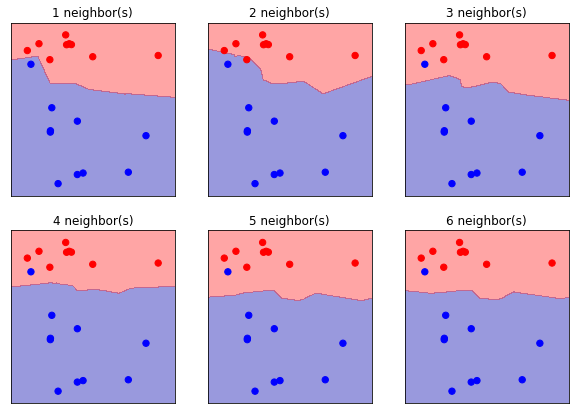
from universe import *
# Load Data
X, y = mglearn.datasets.make_forge()
y[7] = 1
print(f"X[:5] = {X[:5]}")
print(f"y = {y}")
# Train/Test Set Split
forge_X_train, forge_X_test, forge_y_train, forge_y_test = \
train_test_split(X, y, random_state=0)
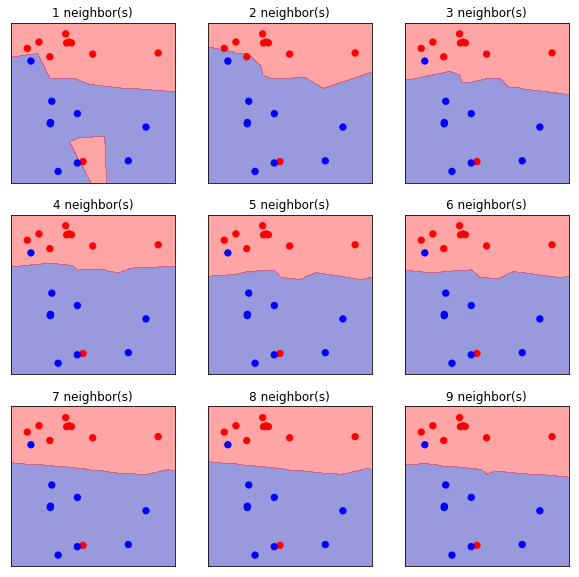
# Train Models and Plot Figures
plt.figure(figsize=(10, 10))
for k in range(1, 10):
plt.subplot(3, 3, k)
# Train Model
clf = KNeighborsClassifier(n_neighbors=k)
clf.fit(forge_X_train, forge_y_train)
# Plot Figure
cmap = mpl.colors.ListedColormap(['b', 'r'])
mglearn.plots.plot_2d_separator(clf, forge_X_train, fill=True, eps=0.5, alpha=.4)
plt.scatter(forge_X_train[:, 0], forge_X_train[:, 1], c=forge_y_train,
cmap=cmap, s=40, lw=1)
plt.title(f"{k} neighbor(s)")
plt.show()
kNN for Regression¶


Linear & Logistic Regression¶
Linear Regression and Logistic Regression (which indeed is a Classification problem) were introduced in previous modules.
Ridge Regularization¶
Cost function¶
Cost function ($k$ features, $n$ samples):
$$ \underset{w_1,\dots,w_k,b}{min\,} \frac{1}{2n} \sum\limits_{i}^{n} (y_\textit{prediction} - y_\textit{ground truth})^2 + \alpha (w_1^2 + \dots + w_k^2 + b^2) $$
Regularization and Parameters¶
from universe import *
# Load and Split Data
dataset = load_boston()
X, y = dataset['data'], dataset['target']
feature_names = dataset['feature_names']
X_train, X_test, y_train, y_test = train_test_split(X, y, random_state=42)
# Train Model and Compute Results
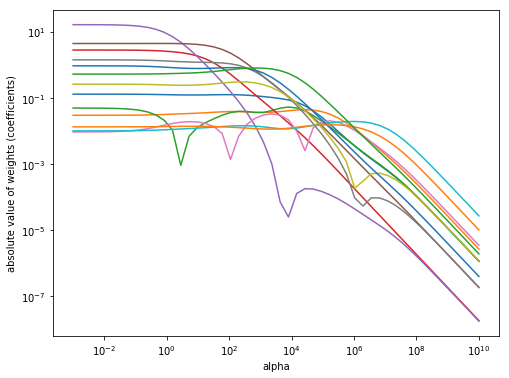
def compute_coefs(alpha):
model = Ridge(alpha=alpha)
model.fit(X_train, y_train)
return model.coef_
compute_coefs = np.vectorize(compute_coefs, signature='()->(n)')
alpha = np.logspace(-3, 10)
coefs = compute_coefs(alpha)
# Plot Figure
plt.figure(figsize=(8, 6))
plt.plot(alpha, np.abs(coefs))
plt.xscale('log')
plt.xlabel('alpha')
plt.ylabel('absolute value of weights (coefficients)')
plt.yscale('log')
plt.show()
Neural Networks¶
Neuron¶
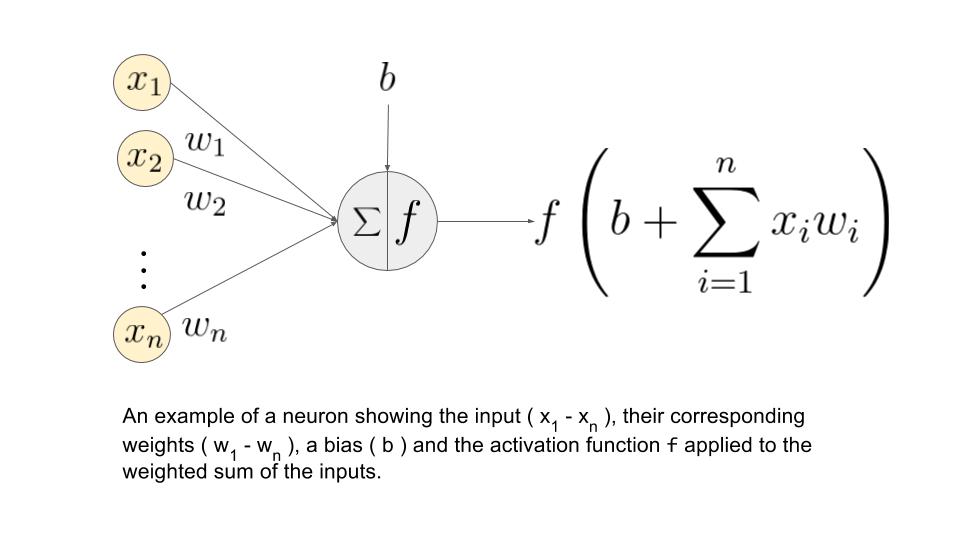
Neural Network¶

Activation Functions¶
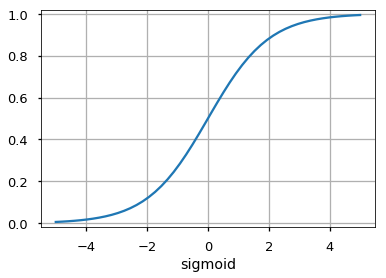
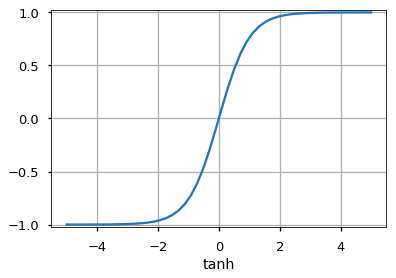
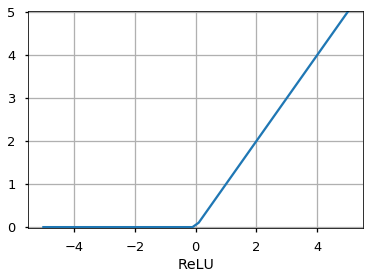
Number of Neurons¶
# Toy Dataset
X, y = make_moons(n_samples=100, noise=0.25, random_state=3)
X_train, X_test, y_train, y_test = train_test_split(
X, y, stratify=y, random_state=42)
#
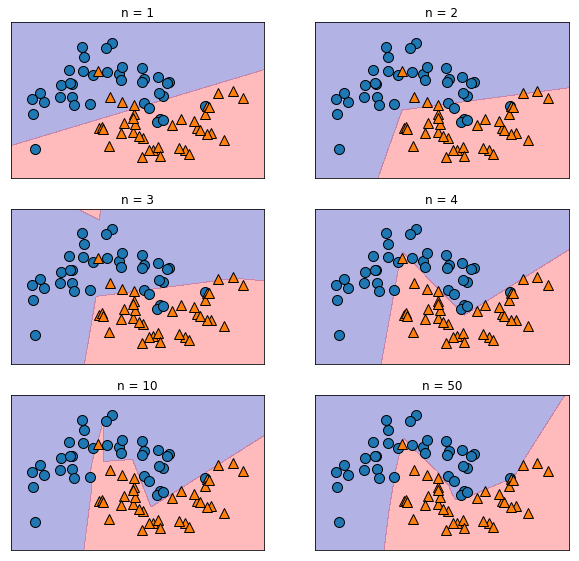
plt.figure(figsize=(10,20))
i = 1
for n in [1, 2, 3, 4, 10, 50]:
mlp = MLPClassifier(solver='lbfgs', activation='relu',
random_state=0, hidden_layer_sizes=[n])
mlp.fit(X_train, y_train)
plt.subplot(6, 2, i)
plt.title(f"n = {n}")
mglearn.plots.plot_2d_separator(mlp, X_train, fill=True, alpha=.3)
mglearn.discrete_scatter(X_train[:, 0], X_train[:, 1], y_train)
i += 1
plt.show()
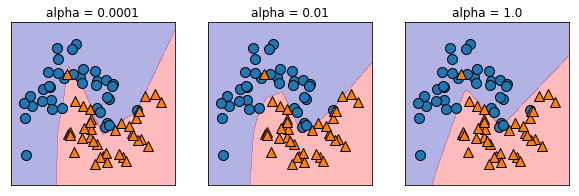
Regularization¶
plt.figure(figsize=(10, 3))
for i, alpha in enumerate([0.0001, 0.01, 1.0]):
mlp = MLPClassifier(solver='lbfgs', random_state=0,
hidden_layer_sizes=[20, 20], alpha=alpha)
mlp.fit(X_train, y_train)
plt.subplot(1, 3, i+1)
mglearn.plots.plot_2d_separator(mlp, X_train, fill=True, alpha=.3)
mglearn.discrete_scatter(X_train[:, 0], X_train[:, 1], y_train)
plt.title(f"alpha = {alpha}")
plt.show()
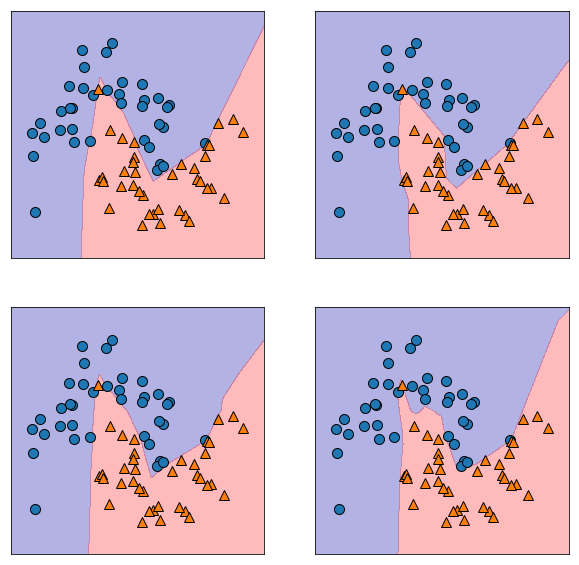
Randomness¶
plt.figure(figsize=(10, 10))
for i in range(4):
plt.subplot(2, 2, i+1)
mlp = MLPClassifier(solver='lbfgs', random_state=i,
hidden_layer_sizes=[20, 20])
mlp.fit(X_train, y_train)
mglearn.plots.plot_2d_separator(mlp, X_train, fill=True, alpha=.3)
mglearn.discrete_scatter(X_train[:, 0], X_train[:, 1], y_train)
plt.show()
Decision Trees¶
Controlling Tree Complexity¶
from universe import *
cancer = load_breast_cancer()
X_train, X_test, y_train, y_test = train_test_split(
cancer.data, cancer.target, stratify=cancer.target, random_state=42)
tree = DecisionTreeClassifier(random_state=0)
tree.fit(X_train, y_train)
training_score = tree.score(X_train, y_train)
test_score = tree.score(X_test, y_test)
print("WITHOUT PRE-PRUNNING")
print(f"Accuracy on training set: {training_score:.3f}")
print(f"Accuracy on test set: {test_score:.3f}")
tree = DecisionTreeClassifier(max_depth=2, random_state=0)
tree.fit(X_train, y_train)
training_score = tree.score(X_train, y_train)
test_score = tree.score(X_test, y_test)
print("")
print("WITH PRE-PRUNNING")
print(f"Accuracy on training set: {training_score:.3f}")
print(f"Accuracy on test set: {test_score:.3f}")
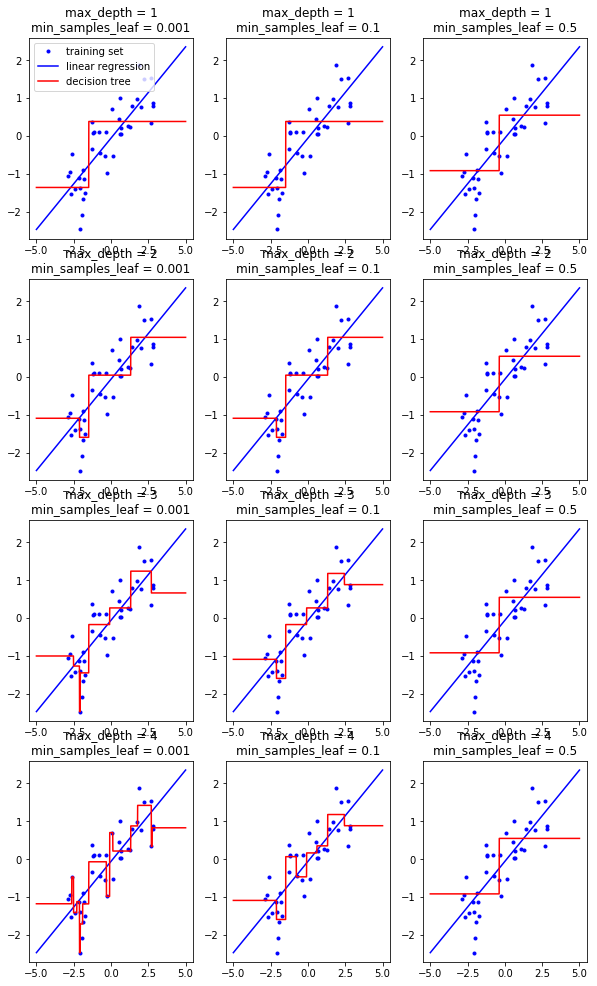
Decision Tree for Regression¶
from universe import *
X, y = mglearn.datasets.make_wave(n_samples=40)
### Exercise: Plot the Figure
# Hint:
'''
i = 1
for max_depth in [1, 2, 3, 4]:
for min_samples_leaf in [0.001, 0.1, 0.5]:
plt.subplot(4, 3, i)
tree = DecisionTreeRegressor(
max_depth=max_depth,
min_samples_leaf=min_samples_leaf)
...
i += 1
plt.show()
''';
Random Forests¶
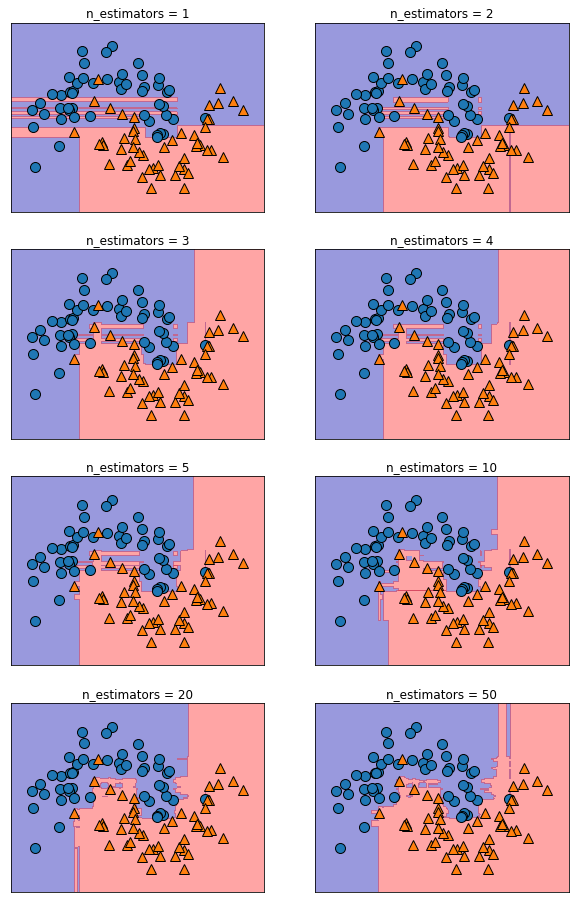
Random Forests¶
from universe import *
X, y = make_moons(n_samples=100, noise=0.25, random_state=3)
i = 1
plt.figure(figsize=(10, 16))
for n_estimators in [1, 2, 3, 4, 5, 10, 20, 50]:
forest = RandomForestClassifier(n_estimators=n_estimators, random_state=2)
forest.fit(X, y);
plt.subplot(4, 2, i)
plt.title(f"n_estimators = {n_estimators}")
mglearn.plots.plot_2d_separator(forest, X, fill=True,
alpha=.4)
mglearn.discrete_scatter(X[:, 0], X[:, 1], y)
i += 1
plt.show()
forest = RandomForestClassifier(max_leaf_nodes=4, n_estimators=3, random_state=2)
forest.fit(X, y)
tree1, tree2, tree3 = forest.estimators_
def display_tree(tree, feature_names):
dot_data = export_graphviz(
tree, out_file=None,
feature_names=feature_names, label='root',
filled=True, impurity=False)
display(graphviz.Source(dot_data))
display_tree(tree1, feature_names=['X[0]', 'X[1]'])
display_tree(tree2, feature_names=['X[0]', 'X[1]'])
display_tree(tree3, feature_names=['X[0]', 'X[1]'])
X_test = np.array([[2, 3]])
print(tree1.predict(X_test))
print(tree2.predict(X_test))
print(tree3.predict(X_test))
print(forest.predict(X_test))
Classification¶
from universe import *
%matplotlib inline
Logistic Regression¶
Odds Ratio¶
$$ Odds=\frac{p}{1-p}$$
$$ p=\frac{Odds}{1+Odds} $$
Assume that probability is in $(0, 1)$ range, excluding 0 and 1.
=> Odds Ratio is in $(0,+\infty)$ range.
=> Logarithm of Odds Ratio is in $(-\infty, +\infty)$ range.
Logit function is defined as logarithm of odds ratio. We model it with a linear combination:
$$ \operatorname{logit}(p)=\ln\left(\frac{p}{1-p}\right) = w_1 x_{1} + \cdots + w_k x_{k} + b$$
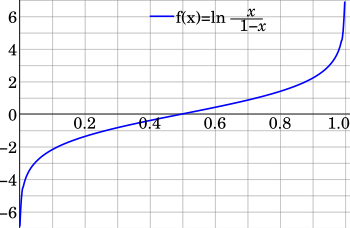
We can compute the probability back from its logit. This is called Logistic Function:
$$ p =\frac{e^{\operatorname{logit}(p)}}{1+e^{\operatorname{logit}(p)}} =\frac{1}{1+e^{-\operatorname{logit}(p)}} =\frac{1}{1+e^{-(w_1 x_{1} + \cdots + w_k x_{k} + c)}} $$

Cost Function¶
L2 Regularization:
$$\underset{w, c}{min\,} \frac{1}{2}\left(w_1^2+\cdots+w_k^2\right) + C \sum_{i=1}^n \ln\left(1 + e^{- y_i (w_1 x_{i,1} + \cdots + w_k x_{i,k} + c)} \right)$$
Binary Classification¶
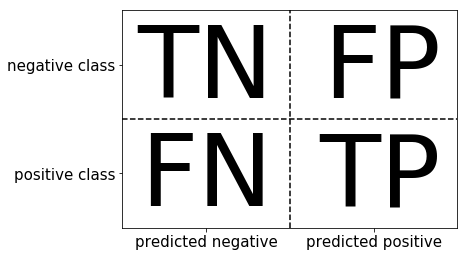
Confusion Matrix¶

Beyond Accuracy¶
Consider a test that can be positive or negative. This test tells you if somebody is drug user or not.
Accuracy -- how often we get correct result (doesn't matter if it's negative or positive)?
$$Accuracy = \frac{TP + TN}{TP + TN + FP + FN}$$
Precision, positive predictive value (PPV) -- when test is positive, how likely it's that one is drug user?
$$Precision = \frac{TP}{TP + FP}$$
Recall, sensitivity, hit rate, true positive rate (TPR) -- when one is drug user, how likely it is that the test will detect it, that is it's positive? $$Recall = \frac{TP}{TP + FN}$$
$F_1$-score -- a metric combining both Precision and Recall.
$$F = 2 \cdot \frac{precision \cdot recall}{precision + recall} = \frac{2}{\frac{1}{precision}+\frac{1}{recall}}$$
Polish names:
- Accuracy = dokładność
- Precision = precyzja
- Recall = czułość
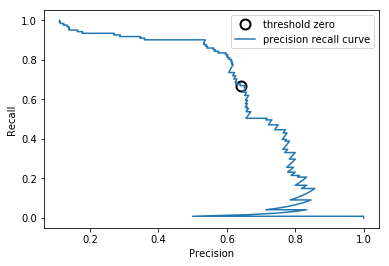
Precision-Recall Curve¶
from universe import *
# Load Data and Split It
X, y = mglearn_make_blobs(n_samples=(4000, 500), centers=2, cluster_std=[7.0, 2],
random_state=22)
X_train, X_test, y_train, y_test = train_test_split(X, y, random_state=0)
# Build And Train Model
svc = SVC(gamma=.05).fit(X_train, y_train)
# Compute Precision-Recall Curve
precision, recall, thresholds = precision_recall_curve(
y_test, svc.decision_function(X_test))
# Find threshold closest to zero
close_zero = np.argmin(np.abs(thresholds))
# Plot Precision-Recall Curve
plt.plot(precision[close_zero], recall[close_zero], 'o', markersize=10,
label="threshold zero", fillstyle="none", c='k', mew=2)
plt.plot(precision, recall, label="precision recall curve")
plt.xlabel("Precision")
plt.ylabel("Recall")
plt.legend()
plt.show()
Usage with Grid Search¶
grid = GridSearchCV(
pipeline,
param_grid,
cv=KFold(n_split=5),
refit='f1_score',
return_train_score=True,
iid=False,
)